|
|
Gaussian Mixture Models (GMMs) are a type of probabilistic model used for clustering and density estimation. They assist in representing data as a combination of different Gaussian distributions, where each distribution is characterized by its mean, covariance, and weight. The Gaussian mixture model probability distribution class allows us to estimate the parameters of such a mixture distribution. In this article, we’ll delve into four types of covariances with GMM models. GMM in Scikit-LearnIn Scikit-Learn, the GaussianMixture class is used for GMM-based clustering and density estimation. The covariances within GMMs play a vital role in shaping the individual Gaussian components of the mixture. GaussianMixture includes several parameters such as n_components, covariance_type, tol, reg_covar, max_iter, n_init, init_params, weights_init, means_init, precisions_init, random_state, warm_start, verbose, and verbose_interval. While we won’t discuss all of these in detail within this article to keep it concise, we will explore them in a separate article. For now, let’s focus on understanding the covariance_type parameter more thoroughly. Understanding and selecting the appropriate covariance type is an important aspect of utilizing GMMs effectively for tasks such as clustering and density estimation. It involves considerations of the inherent structure and relationships within the data. Covariance Types in Gaussian MixtureIn Gaussian Mixture Models, there are four covariance types available:
These covariance types in Gaussian Mixture offer flexibility in modeling the distribution of data. Working with GMM Covariances in Scikit-LearnTo work with GMM covariances in Scikit-Learn, let’s delve deeper into the model with in-built wine dataset. Step 1: Importing Required LibrairesThe very first step is to import the necessary libraries. When working with GMM covariances in Scikit-Learn, it’s essential to have the Scikit-Learn library. An important consideration is to ensure that the library is already installed in your Python environment to avoid any import errors. Here, we specifically need the “GaussianMixture” module from Scikit-Learn, so we will import Scikit-Learn. Additionally, we will import the NumPy library to facilitate data manipulation. To import these libraries, you can use the following code: Python3
Step 2: Data PreparationData preparation is a crucial step in our program. Ensuring that our data is in the correct format for GMM is essential. We must prepare our data accordingly. To begin, let’s load wine dataset for this task, we will leverage the NumPy library. Python3
Step 3: Initializing Gaussian Mixture Model:In this step, we will initialize a Gaussian Mixture Model. To do this, we specify two key parameters:
To work with Gaussian Mixture in Scikit-Learn, we will use the Python3
Step 4: Fitting the GMM Model:In this step, we will fit our GMM model using our prepared data. This fitting process will estimate the model’s parameters, including the specified covariance type, along with other essential components. Fitting the GMM model is a crucial step that helps us understand the underlying structure of the data and how it relates to the chosen covariance type. Python3
Step 5: Accessing CovariancesYou can access the covariance matrices of the components through the “covariances_” attribute of our fitted GMM model. The shape of these covariance matrices depends on the specified ‘covariance_type‘. This will help accessing the covariances. Python3
Step 6: Using the GMM Model for Clustering or PredictionsWith our GMM model fully prepared, the final step is to utilize the model for clustering or making predictions, depending on the specific task at hand. Python3
Step 7 : VisualizationsPython3
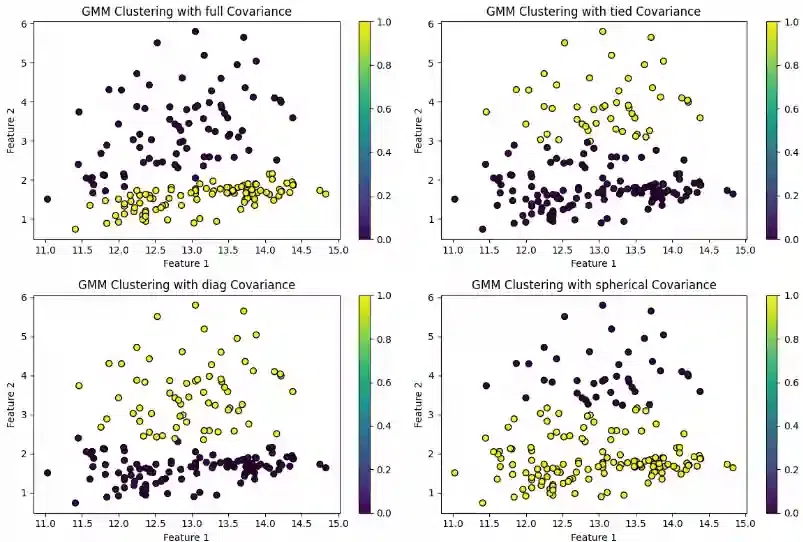
Output: Covariance Matrix (full - Component):  Gaussian Mixture Model All four plots show the relationship between the number of clusters and the BIC score.
Overall, the choice of covariance structure can have a significant impact on the performance of GMM clustering. It is important to experiment with different options to find the one that best fits the data. Choosing an appropriate covariance type is often task-dependent. For datasets with varying shapes and orientations, ‘full’ covariance might be more suitable. In cases where components are expected to have similar shapes, ‘tied’ or ‘spherical’ covariance might be more appropriate. ConclusionGaussian Mixture Models offer flexible clustering with diverse covariance structures. Model selection, especially covariance type, impacts performance; choose wisely for optimal results. |
Reffered: https://www.geeksforgeeks.org
| AI ML DS |
Type: | Geek |
Category: | Coding |
Sub Category: | Tutorial |
Uploaded by: | Admin |
Views: | 13 |